Radar backscatter RGB composites
Since radar data is quite complex to interpret, temporal RGB composites are a nice way to vividly display changes in backscatter over time. yeoda is the perfect tool to assist in this use case, since it allows to select and filter data in an interactive and flexible manner. As input we use radiometric terrain-flattened gamma nought data in VV polarisation produced by the GEO Department of TU Wien within the frame of the ACube project. To limit the necessary disk space, this benchmark dataset consists of data from July 2016, July 2018, and July 2021 for a region aroung Lake Neusiedl, Austria. Our aim is now to compute monthly averages for these three years and visualise them as an RGB composite.
Lets start! First, we need to collect all file paths,
[2]:
import os
import glob
ds_path = r"D:\data\code\yeoda\2022_08__docs\radar_data"
filepaths = glob.glob(os.path.join(ds_path, "*.tif"))
filepaths[:10]
[2]:
['D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160702_163408--_GMR------_S1AIWGRDH1VVA_175_A0105_EU010M_E053N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160704_050935--_GMR------_S1AIWGRDH1VVD_022_A0105_EU010M_E052N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160704_050935--_GMR------_S1AIWGRDH1VVD_022_A0105_EU010M_E053N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160704_051000--_GMR------_S1AIWGRDH1VVD_022_A0105_EU010M_E052N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160706_045311--_GMR------_S1AIWGRDH1VVD_051_A0105_EU010M_E053N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160706_045336--_GMR------_S1AIWGRDH1VVD_051_A0105_EU010M_E053N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160707_164202--_GMR------_S1AIWGRDH1VVA_073_A0105_EU010M_E052N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160707_164202--_GMR------_S1AIWGRDH1VVA_073_A0105_EU010M_E053N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160707_164227--_GMR------_S1AIWGRDH1VVA_073_A0105_EU010M_E052N015T1.tif',
'D:\\data\\code\\yeoda\\2022_08__docs\\radar_data\\D20160707_164227--_GMR------_S1AIWGRDH1VVA_073_A0105_EU010M_E053N015T1.tif']
define the dimensions we are interested in and create a DataCubeReader instance. The naming convention of ACube data follows the SgrtFilename convention, which is already defined in geopathfinder.
[3]:
from yeoda.datacube import DataCubeReader
from geopathfinder.naming_conventions.sgrt_naming import SgrtFilename
dimensions = ['time', 'tile_name', 'relative_orbit']
dc_reader = DataCubeReader.from_filepaths(filepaths, fn_class=SgrtFilename, dimensions=dimensions,
stack_dimension="time", tile_dimension="tile_name")
dc_reader
[3]:
DataCubeReader -> GeoTiffReader(time, MosaicGeometry):
filepath tile_name \
0 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
1 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
2 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
3 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
4 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
.. ... ...
207 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
208 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
209 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
210 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
211 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
relative_orbit time
0 175 2016-07-02 16:34:08
1 22 2016-07-04 05:09:35
2 22 2016-07-04 05:09:35
3 22 2016-07-04 05:10:00
4 51 2016-07-06 04:53:11
.. ... ...
207 73 2021-07-29 16:42:42
208 73 2021-07-29 16:43:07
209 73 2021-07-29 16:43:07
210 175 2021-07-30 16:33:43
211 175 2021-07-30 16:34:08
[212 rows x 4 columns]
To stay within a reasonable time when computing temporal averages, we can pre-filter the datacube per relative orbit first. An appropriate selection would be for instance to take the orbit with the largest file sizes, i.e. containing the largest amount of measurements. To do so, we can add a new dimension to the datacube containing the size in MB for each file.
[4]:
file_sizes = [int(os.path.getsize(filepath)/1e6) for filepath in dc_reader['filepath']]
dc_reader.add_dimension("file_size", file_sizes, inplace=True)
[4]:
DataCubeReader -> GeoTiffReader(time, MosaicGeometry):
filepath tile_name \
0 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
1 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
2 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
3 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
4 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
.. ... ...
207 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
208 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
209 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
210 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
211 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E053N015T1
relative_orbit time file_size
0 175 2016-07-02 16:34:08 95
1 22 2016-07-04 05:09:35 24
2 22 2016-07-04 05:09:35 3
3 22 2016-07-04 05:10:00 144
4 51 2016-07-06 04:53:11 14
.. ... ... ...
207 73 2021-07-29 16:42:42 127
208 73 2021-07-29 16:43:07 77
209 73 2021-07-29 16:43:07 64
210 175 2021-07-30 16:33:43 18
211 175 2021-07-30 16:34:08 79
[212 rows x 5 columns]
Now we need to look at each tile separately to check if the relative orbit covers both tiles sufficiently.
[5]:
dc_reader.select_tiles(["E052N015T1"]).sort_by_dimension("file_size").file_register
[5]:
| filepath | tile_name | relative_orbit | time | file_size | |
|---|---|---|---|---|---|
| 74 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-13 05:02:17 | 4 |
| 90 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-19 05:01:35 | 4 |
| 57 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-07 05:01:34 | 4 |
| 107 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-25 05:02:17 | 4 |
| 123 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-31 05:01:36 | 4 |
| ... | ... | ... | ... | ... | ... |
| 105 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-25 05:01:52 | 184 |
| 42 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-01 05:01:51 | 184 |
| 121 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-31 05:01:11 | 185 |
| 88 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-19 05:01:10 | 185 |
| 200 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-27 05:01:34 | 187 |
96 rows × 5 columns
[6]:
dc_reader.select_tiles(["E053N015T1"]).sort_by_dimension("file_size").file_register
[6]:
| filepath | tile_name | relative_orbit | time | file_size | |
|---|---|---|---|---|---|
| 70 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 22 | 2018-07-12 05:09:10 | 3 |
| 161 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 22 | 2021-07-14 05:10:11 | 3 |
| 190 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 146 | 2021-07-22 16:51:10 | 3 |
| 188 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 146 | 2021-07-22 16:50:45 | 3 |
| 47 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 146 | 2018-07-02 16:50:25 | 3 |
| ... | ... | ... | ... | ... | ... |
| 89 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-19 05:01:10 | 186 |
| 122 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-31 05:01:11 | 187 |
| 106 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-25 05:01:52 | 187 |
| 201 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-27 05:01:34 | 188 |
| 164 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-15 05:01:27 | 189 |
116 rows × 5 columns
It looks like relative orbit number 124 is a good choice. Thus, we select this relative orbit number in the datacube
[7]:
dc_reader.select_by_dimension(lambda r: r == 124, name='relative_orbit', inplace=True)
dc_reader.file_register
[7]:
| filepath | tile_name | relative_orbit | time | file_size | |
|---|---|---|---|---|---|
| 10 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2016-07-11 05:01:25 | 23 |
| 11 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2016-07-11 05:01:25 | 75 |
| 12 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2016-07-11 05:01:50 | 167 |
| 13 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2016-07-11 05:01:50 | 119 |
| 26 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2016-07-23 05:01:36 | 149 |
| 27 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2016-07-23 05:01:36 | 183 |
| 28 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2016-07-23 05:02:01 | 38 |
| 29 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2016-07-23 05:02:01 | 4 |
| 42 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-01 05:01:51 | 184 |
| 43 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-01 05:01:51 | 186 |
| 44 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-01 05:02:16 | 4 |
| 55 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-07 05:01:09 | 183 |
| 56 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-07 05:01:09 | 184 |
| 57 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-07 05:01:34 | 4 |
| 72 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-13 05:01:52 | 184 |
| 73 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-13 05:01:52 | 186 |
| 74 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-13 05:02:17 | 4 |
| 88 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-19 05:01:10 | 185 |
| 89 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-19 05:01:10 | 186 |
| 90 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-19 05:01:35 | 4 |
| 105 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-25 05:01:52 | 184 |
| 106 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-25 05:01:52 | 187 |
| 107 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-25 05:02:17 | 4 |
| 121 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-31 05:01:11 | 185 |
| 122 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2018-07-31 05:01:11 | 187 |
| 123 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2018-07-31 05:01:36 | 4 |
| 127 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-03 05:01:18 | 73 |
| 128 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-03 05:01:18 | 125 |
| 129 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-03 05:01:43 | 119 |
| 130 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-03 05:01:43 | 69 |
| 146 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-09 05:02:03 | 118 |
| 147 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-09 05:02:03 | 172 |
| 148 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-09 05:02:28 | 75 |
| 149 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-09 05:02:28 | 24 |
| 163 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-15 05:01:27 | 176 |
| 164 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-15 05:01:27 | 189 |
| 165 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-15 05:01:52 | 15 |
| 181 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-21 05:02:04 | 116 |
| 182 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-21 05:02:04 | 167 |
| 183 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-21 05:02:29 | 74 |
| 184 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-21 05:02:29 | 24 |
| 199 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-27 05:01:09 | 4 |
| 200 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E052N015T1 | 124 | 2021-07-27 05:01:34 | 187 |
| 201 | D:\data\code\yeoda\2022_08__docs\radar_data\D2... | E053N015T1 | 124 | 2021-07-27 05:01:34 | 188 |
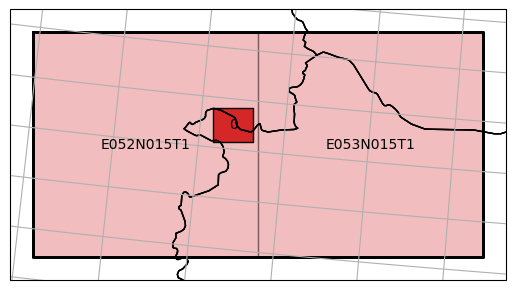
define a bounding box covering the southern part of Lake Neusiedl,
[8]:
bbox = [(5280269, 1551367), (5297621, 1566199)]
dc_reader.select_bbox(bbox, inplace=True)
plot_extent = dc_reader.mosaic.parent_root.outer_extent
extent_bfr = 10e3
plot_extent = [plot_extent[0] - extent_bfr, plot_extent[1] - extent_bfr,
plot_extent[2] + extent_bfr, plot_extent[3] + extent_bfr]
ax = dc_reader.mosaic.parent_root.plot(label_tiles=True, extent=plot_extent, alpha=0.3)
dc_reader.mosaic.plot(label_tiles=True, ax=ax, extent=plot_extent)
[8]:
<GeoAxesSubplot:>
split the data temporally to have a datacube for each month/year,
[9]:
dc_months = dc_reader.split_by_temporal_freq('Y')
dc_months[0]
[9]:
DataCubeReader -> GeoTiffReader(time, MosaicGeometry):
filepath tile_name \
10 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
12 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
26 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
28 D:\data\code\yeoda\2022_08__docs\radar_data\D2... E052N015T1
relative_orbit time file_size
10 124 2016-07-11 05:01:25 23
12 124 2016-07-11 05:01:50 167
26 124 2016-07-23 05:01:36 149
28 124 2016-07-23 05:02:01 38
and finally read the data, decode it, convert between dB and linear units and vice versa, apply a mean operation, and store it in an array representing the RGB composite. For performing decoding and unit conversion, we need to create some helper functions before.
[10]:
def gamma_rtf_decoder(ds):
ar = ds['Gamma_RTF'].data
ar = ar.astype(float)
ar[ar == -9999] = np.nan
ar = ar * 0.01
return ar
def lin2db(ar):
return 10. * np.log10(ar)
def db2lin(ar):
return 10. ** (ar / 10.)
[11]:
import numpy as np
import xarray as xr
rgb_comp = np.zeros((*dc_reader.mosaic.tiles[0].shape, 3))
for i, dc_month in enumerate(dc_months):
dc_month.read(band_names='Gamma_RTF')
dc_month.apply_nan()
gamma_rtf = gamma_rtf_decoder(dc_month.data_view)
rgb_layer = lin2db(np.nanmean(db2lin(gamma_rtf), axis=0))
rgb_comp[..., i] = rgb_layer
Before being able to visualise it with plt.imshow(), we need to convert dB values to RGB values in the range (0, 1). Thus, we create a function for linearly scaling the values
[12]:
def linear_scale_range(x, min_x, max_x, min_y, max_y):
return ((x - min_x) / (max_x - min_x)) * (max_y - min_y) + min_y
and apply to each layer with a fine-tuned input range.
[13]:
rgb_comp[..., 0] = linear_scale_range(rgb_comp[..., 0], -22, -5, 0, 1)
rgb_comp[..., 1] = linear_scale_range(rgb_comp[..., 1], -20, -6, 0, 1)
rgb_comp[..., 2] = linear_scale_range(rgb_comp[..., 2], -21, -6, 0, 1)
The current setup would have the following mapping (R, G, B) -> (2016, 2018, 2021). To highlight recent changes in red, we can simply flip the array
[14]:
rgb_comp = np.flip(rgb_comp, axis=2)
and pass it to plt.imshow().
[15]:
import matplotlib.pyplot as plt
plt.imshow(rgb_comp)
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
[15]:
<matplotlib.image.AxesImage at 0x1a3257e3610>
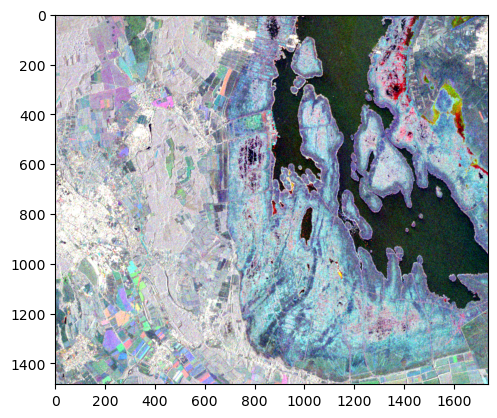
Besides a colourful representation of agricultural areas due to crop rotation, changes of the vegetation state of the reed belt (blueish border line around the water surface) and a dry-out of smaller ponds/swamps in the upper-right corner are clearly visible.