Loading SCATSAR soil water index (SWI)
This example is about loading time series data from the so-called SCATSAR (soil water index) SWI dataset produced by the GEO Department at TU Wien. The (deprecated) dataset consists of daily NetCDF files being located/tiled in the Equi7Grid system. Each file contains encoded SWI data for different temporal lags (“T”) and some quality flag layers.
The first step is to collect all file paths.
[2]:
import os
import glob
import numpy as np
ds_path = r"D:\data\code\yeoda\2022_08__docs\scatsarswi"
filepaths = glob.glob(os.path.join(ds_path, "*.nc"))
filepaths[:10]
[2]:
['D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190101_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N012T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190101_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N018T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190102_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N012T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190102_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N018T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190103_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N012T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190103_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N018T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190104_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N012T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190104_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N018T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190105_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N012T6.nc',
'D:\\data\\code\\yeoda\\2022_08__docs\\scatsarswi\\M20190105_120000--_SWI------_SCATSAR-55VV-_---_C0418_EU500M_E048N018T6.nc']
Unfortunately, the NetCDF files do not comply with the latest CF conventions needed for rioxarray, which is used to retrieve the geospatial information from a file. Therefore, we need to set the mosaic and its CRS manually, which is a quick job since the exemplary dataset only consists of two tiles.
[3]:
from geospade.crs import SpatialRef
from geospade.raster import Tile, MosaicGeometry
sref_wkt = 'PROJCS["Azimuthal_Equidistant",GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",' \
'SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],' \
'UNIT["Degree",0.017453292519943295]],PROJECTION["Azimuthal_Equidistant"],' \
'PARAMETER["false_easting",5837287.81977],PARAMETER["false_northing",2121415.69617],' \
'PARAMETER["central_meridian",24.0],PARAMETER["latitude_of_origin",53.0],UNIT["Meter",1.0]]"'
sref = SpatialRef(sref_wkt)
n_rows, n_cols = 1200, 1200
tiles = [Tile(n_rows, n_cols, sref, geotrans=(48e5, 500, 0, 24e5, 0, -500), name="E048N018T6"),
Tile(n_rows, n_cols, sref, geotrans=(48e5, 500, 0, 18e5, 0, -500), name="E048N012T6")]
mosaic = MosaicGeometry.from_tile_list(tiles)
As a final preparations step, we specify the appropriate naming convention - in this case there is already an existing one, SgrtFilename- and use xarray to retrieve information from the files.
[4]:
from yeoda.datacube import DataCubeReader
from veranda.raster.native.netcdf import NetCdfXrFile
from geopathfinder.naming_conventions.sgrt_naming import SgrtFilename
dimensions = ['dtime_1', 'tile_name']
dc_reader = DataCubeReader.from_filepaths(filepaths, fn_class=SgrtFilename, dimensions=dimensions,
stack_dimension="dtime_1", tile_dimension="tile_name",
mosaic=mosaic,
file_class=NetCdfXrFile)
dc_reader
[4]:
DataCubeReader -> NetCdfReader(dtime_1, MosaicGeometry):
filepath tile_name dtime_1
0 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N012T6 2019-01-01
1 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N018T6 2019-01-01
2 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N012T6 2019-01-02
3 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N018T6 2019-01-02
4 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N012T6 2019-01-03
.. ... ... ...
725 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N018T6 2019-12-29
726 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N012T6 2019-12-30
727 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N018T6 2019-12-30
728 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N012T6 2019-12-31
729 D:\data\code\yeoda\2022_08__docs\scatsarswi\M2... E048N018T6 2019-12-31
[730 rows x 3 columns]
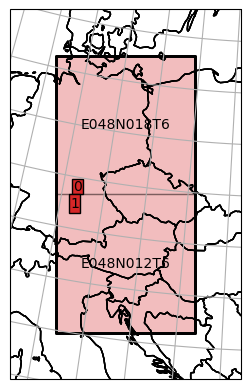
As an example, we can define a bounding box in a different coordinate system to read data from.
[5]:
from geospade.crs import SpatialRef
bbox = [(48.64, 10.61), (49.9, 11.1)]
dc_reader.select_bbox(bbox, sref=SpatialRef(4326), inplace=True)
plot_extent = dc_reader.mosaic.parent_root.outer_extent
extent_bfr = 200e3
plot_extent = [plot_extent[0] - extent_bfr, plot_extent[1] - extent_bfr,
plot_extent[2] + extent_bfr, plot_extent[3] + extent_bfr]
ax = dc_reader.mosaic.parent_root.plot(label_tiles=True, extent=plot_extent, alpha=0.3)
dc_reader.mosaic.plot(label_tiles=True, ax=ax, extent=plot_extent)
[5]:
<GeoAxesSubplot:>
Since the NetCDF files lack in CF compliance, we need to manually specify a decoding function, which basically applies all internal decoding attributes and resets the no data value to 254.
[6]:
import numpy as np
def decoder(dar, scale_factor=1, offset=0, **kwargs):
dar = dar.where(dar != 254, np.nan)
dar = dar * scale_factor + offset
return dar
Finally, we can read data from this bounding box with a set of data variables we are interested in,
[7]:
data_variables = ["SWI_T002", "SWI_T020", "SWI_T100"]
dc_reader.read(engine='xarray', data_variables=data_variables, decoder=decoder, parallel=False)
replace no data values,
[8]:
dc_reader.apply_nan()
dc_reader.data_view
[8]:
<xarray.Dataset>
Dimensions: (x: 121, y: 291, time: 365)
Coordinates:
* x (x) float64 4.855e+06 4.855e+06 ... 4.914e+06 4.915e+06
* y (y) float64 1.865e+06 1.865e+06 ... 1.721e+06 1.72e+06
* time (time) float64 4.346e+04 4.347e+04 ... 4.383e+04 4.383e+04
spatial_ref int32 0
Data variables:
SWI_T002 (time, y, x) float64 nan nan nan nan nan ... nan nan nan nan
SWI_T020 (time, y, x) float64 nan nan nan nan nan ... nan nan nan nan
SWI_T100 (time, y, x) float64 nan nan nan nan nan ... nan nan nan nandefine a point to retrieve a single-location time series from,
[9]:
poi = 48.7, 10.7
dc_ts = dc_reader.select_xy(*poi, sref=SpatialRef(4326))
dc_ts.data_view
[9]:
<xarray.Dataset>
Dimensions: (x: 1, y: 1, time: 365)
Coordinates:
* x (x) float64 4.863e+06
* y (y) float64 1.732e+06
* time (time) float64 4.346e+04 4.347e+04 ... 4.383e+04 4.383e+04
spatial_ref int32 0
Data variables:
SWI_T002 (time, y, x) float64 94.5 92.0 88.5 88.5 ... 89.5 89.5 83.0
SWI_T020 (time, y, x) float64 85.5 85.5 85.5 85.5 ... 84.5 84.5 84.0
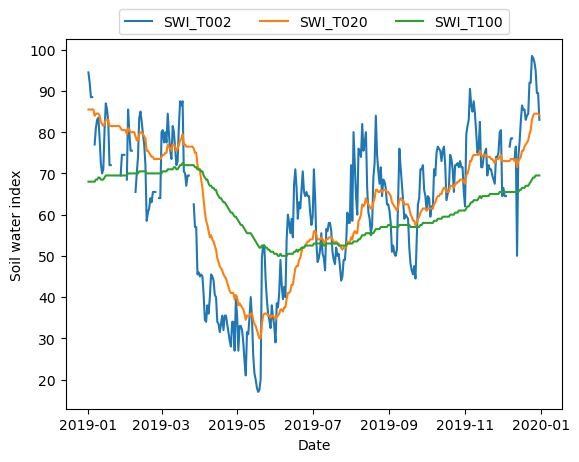
SWI_T100 (time, y, x) float64 68.0 68.0 68.0 68.0 ... 69.5 69.5 69.5and plot it.
[10]:
import matplotlib.pyplot as plt
ts = np.unique(dc_reader['dtime_1'])
plt.plot(ts, dc_ts.data_view['SWI_T002'].data.flatten(), label='SWI_T002')
plt.plot(ts, dc_ts.data_view['SWI_T020'].data.flatten(), label='SWI_T020')
plt.plot(ts, dc_ts.data_view['SWI_T100'].data.flatten(), label='SWI_T100')
plt.xlabel('Date')
plt.ylabel('Soil water index')
_ = plt.legend(ncol=3, loc='upper center', bbox_to_anchor=(0.5, 1.1))